Engineer the world to be a better place!
Some information to be appeared here soon!
Agenda Setup
Aсcording to the official Documentation for beginners the following things are necessary for starting to use Org-mode: Nothing at all!
From my side, I have to add that one probably still needs some
experience in using of Emacs itself. Basic setup to emacs
configuration file for Org-mode (~/.emacs in my case):
(add-to-list 'auto-mode-alist '("\\.org\\'" . org-mode))
(global-set-key "\C-cl" 'org-store-link)
(global-set-key "\C-ca" 'org-agenda)
; JigurdaThe first string will tell emacs automatically go to org-mode when
a file with .org extension is opened. The second string bind C-c+l (big
C in emacs notation is Ctrl button, - to press it simultaneously,
+ to press in a sequence) key for storing a link to a particular
string particular file. This link can be inserted to org-file later
with C-c+C-l. To open the link one can use C-c+C-o. The third string
bind C-c+a for agenda window:
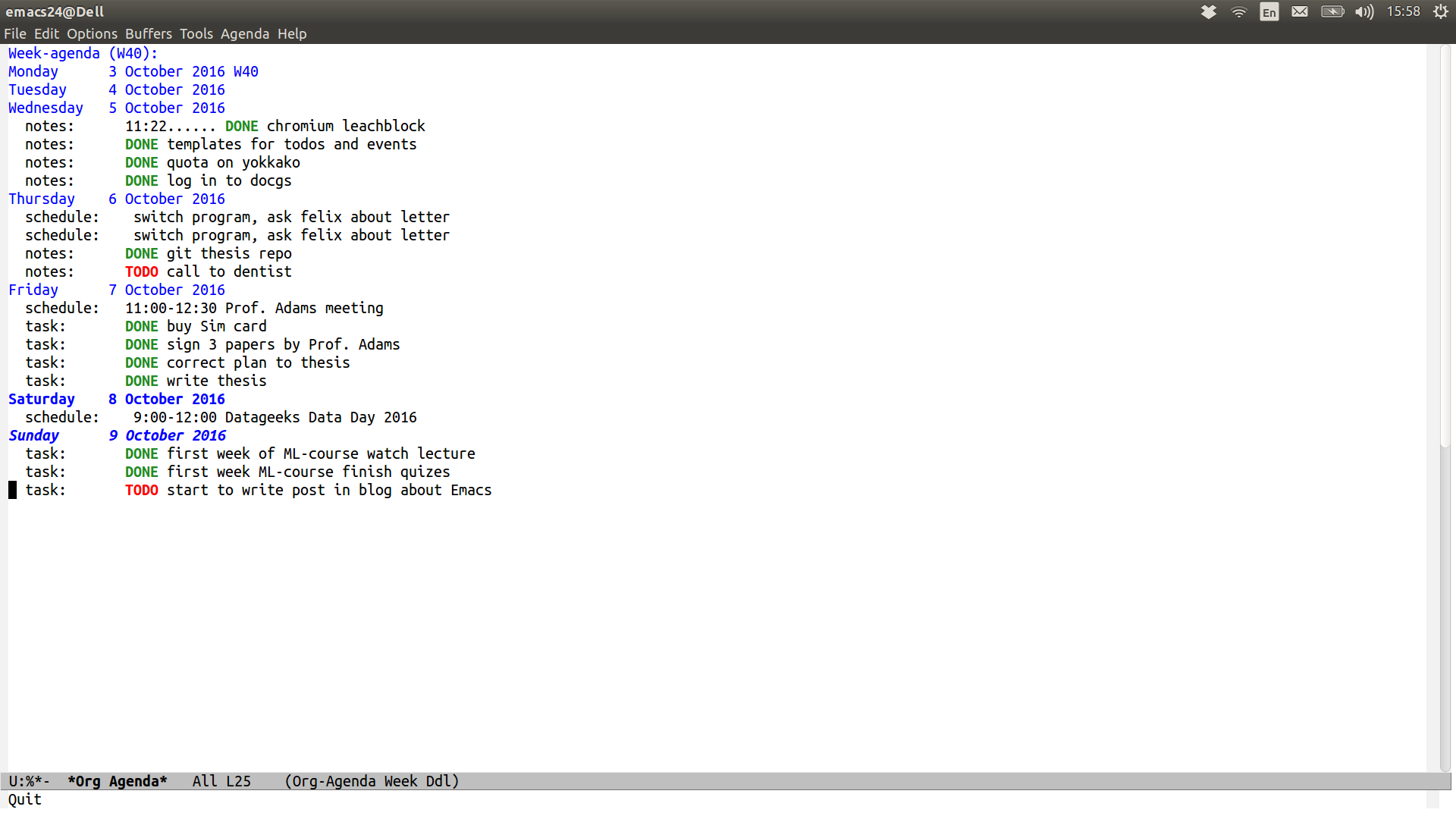
For my Agenda I use two different files: tasks.org and
schedule.org. In schedule.org I store events and task.org is used
for everyday’s TODOs.
(define-key global-map "\C-cc" 'org-capture)
; Log done state in TODOs
(setq org-log-done t)
; Set Org-Capture templates
(setq org-capture-templates
`(
("t" "todo" entry (file+headline "~/Calendars/task.org" "Tasks")
(file "~/org/tasks.orgcaptmpl"))
("e" "event" entry (file+datetree "~/Calendars/schedule.org" "Events")
(file "~/org/events.orgcaptmpl"))
))
(setq org-agenda-files (list "~/Calendars/task.org"
"~/Calendars/schedule.org"
))Org capture available with C-c+c starts dialogue box where I can choose
which type of notes I want to create. With t the task note will be
created. The task event will be stored in the file
~/Calendars/task.org. I use ~/org/tasks.orgcaptmpl as a template
for a task note. The template file for task look like:
** TODO %t %^{Title}
%?%t is time and %^{Title} is the title. Guide to notes template with more
options is available
(http://emacsnyc.org/assets/documents/how-i-use-org-capture-and-stuff.pdf). The
same is for event type of notes.
Google Calendar in Emacs: Calfw
The other usefull tools for Org-mode Agenda are Calfw and Org-Gcal (thanks to http://jameswilliams.be/blog/2016/01/11/Taming-Your-GCal.html).
Calfw allows agenda to look nicely. It is actually like to have Google-Calendar view inside Emacs. For installation it is possible to simply clone emacs-calfw from the repo (https://github.com/kiwanami/emacs-calfw) and add to .emacs file the following strings:
(load "~/emacs/emacs-calfw/calfw.el")
(load "~/emacs/emacs-calfw/calfw-org.el")
(require 'calfw)
(require 'calfw-org)
(defalias 'ca 'cfw:open-org-calendar)Then Calfw view is available with M-x ca:
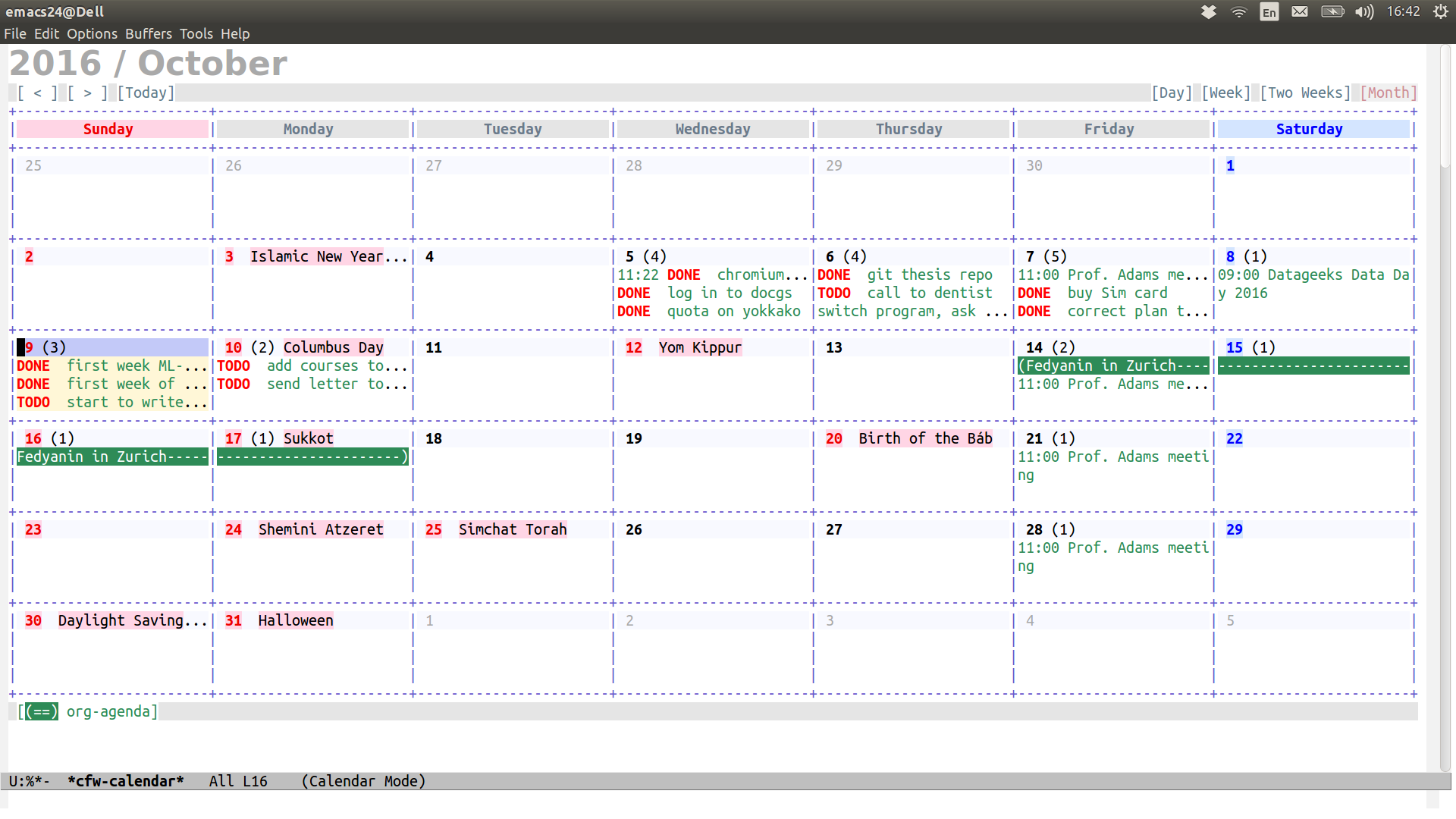
Google Calendar in Emacs: synchronization with Org-Gcal
Org-Gcal is used for synchronyzation with Google-calendar. To install it, one needs to clone:
- https://github.com/tkf/emacs-request
- https://github.com/jwiegley/alert
- https://github.com/kiwanami/emacs-deferred
- https://github.com/myuhe/org-gcal.el
and load to .emacs file:
(load "~/emacs/alert/alert.el")
(load "~/emacs/emacs-request/request.el")
(load "~/emacs/emacs-deferred/deferred.el")
(load "~/emacs/emacs-request/request-deferred.el")
(load "~/emacs/org-gcal.el/org-gcal.el")
(require 'org-gcal)The last thing to set up synchronization is to create application in Google-API that is allowed to access to Google-calendar. The instruction that is provided on https://github.com/myuhe/org-gcal.el seems outdated. Here I list the new up-to-date instruction:
- Go on Google Developers Console
- Create a project (with any name)
- Click on Google APIs
- Choose Calendar API
- Press “Enable” button to enable Calendar API for your project
- Press “Go to credentials” button to set up access
- Choose “OAuth client ID”, First you will offer to setup project consent screen. You need to sepcify only “Product name shown to users” line and press save button.
- After saving, “Create client ID” will appear and you should choose Application type “Other” and “Create”
- save client ID and client secret
- Go to Google Calendar
- Go to settings, Choose “Calendar” menu. Press on the name of calendar you want to synchronize.
- Near ICAL and HTML you will find Calendar ID: callendar id
So you finally have three different things: client ID, client secret and callendar id. Now to set up synchronization add the following strings to your emacs configuration file:
(setq org-gcal-client-id "cliend ID"
org-gcal-client-secret "client secret"
org-gcal-file-alist '(("callendar id" . "~/Calendars/schedule.org")
))TODOs
- It is still not clear for me how to set up synchronization with Google Calendars Tasks.
Back in 2011 I have started my PhD in Technical University of Munich. During my first years, lots of my acquaintances asked me for an advice in searching PhD positions in EU and US as well as what are the best chairs to apply. Though I learned about the best chairs and Universities in Aerodynamics and related fields, I had no idea what to recommend to people who wanted to apply for a PhD position, say, in Chemistry. Still, I wanted to help. I searched for a ranking, which gives you the best people to supervise PhD in the chosen field and did not find any.
So together with my friend, we thought how to create such a ranking. The best idea which came to our minds is to sort people by the number of publications for the last 10-15 years in the chosen field. Such a criterion was imposed due to the idea, that if somebody is so highly productive and publishes a lot of papers, then it would be easier for new PhD students to publish and to graduate with a PhD degree in the field of their interest. In this sense, the number of publications might be more representative, rather than h-index. The criteria of the most numbers of publications might not be accurate to define the true ranking who is the best in the field. After all, they have Nobel prize, Fields medal, Abel prize for such purposes. But the ranking based on the number of publications gives top 10-30 people in the field to supervise PhD thesis. Usually, such highly productive people are heads of chairs, where the graduate student can apply.
We used the web-of-science database and wrote a small script in Python. On the web-of-science search tool one can set different types of filters, such as choosing location, field and years of publications.
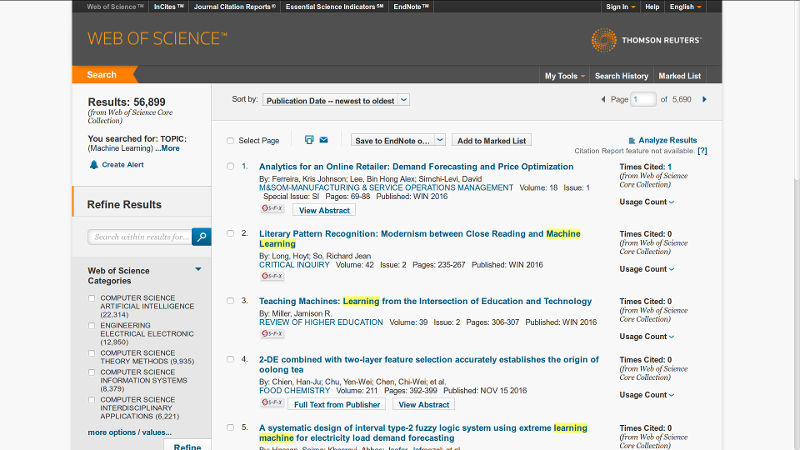
The search results could be downloaded in txt files. The inconvenience is due to WoS allow to download up to 500 records at once, so to search through 10000 different publications one needs to press download button 20 times.
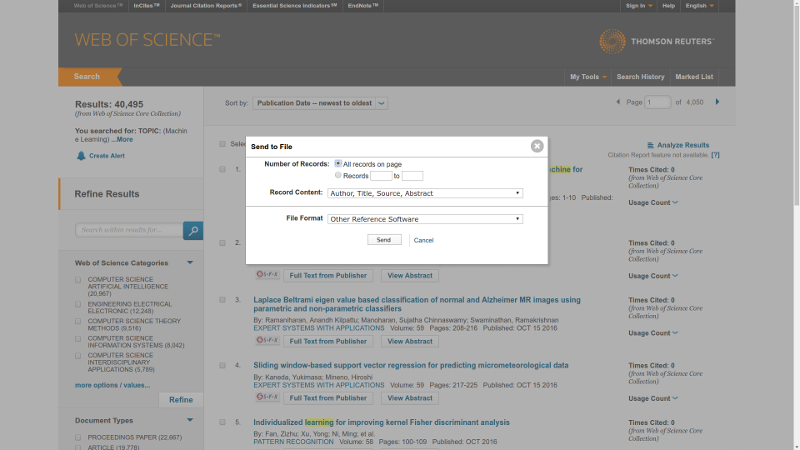
After results are stored in the folder FOLDER, just simply type in command line:
:~$ git clone https://github.com/azarnyx/parse.py
:~$ cd parse.py
:~$ python parse.py -d /Path/To/FOLDERSo I use the script to estimate top-20 people published the most papers that satisfy query “Machine Learning” for 2000-2016 years. I searched through 10000 most cited papers from Web of Science search tool. Here is the result:
| Place | Name | # Publications |
|---|---|---|
| 1. | Herrera, Francisco | 35 |
| 2. | Huang, Guang-Bin | 28 |
| 3. | Mueller, Klaus-Robert | 24 |
| 4. | Zhou, Zhi-Hua | 20 |
| 5. | Wang, Ji-Bo | 17 |
| 6. | Fernandez, Alberto | 17 |
| 7. | Scholkopf, B | 17 |
| 8. | Jennings, NR | 17 |
| 9. | Staab, S | 17 |
| 10. | Bruzzone, Lorenzo | 16 |
| 11. | Pedrycz, Witold | 16 |
| 12. | Herrera, F | 16 |
| 13. | Blankertz, Benjamin | 15 |
| 14. | Polat, Kemal | 15 |
| 15. | Hu, Qinghua | 14 |
| 16. | Suykens, JAK | 14 |
| 17. | Fan, BT | 14 |
| 18. | Han, JW | 14 |
| 19. | Wooldridge, M | 14 |
| 20. | Hu, ZD | 13 |
The alternative to Web Of Science database is to use Google Scholar database or PubMed. Pubmed is mostly related to medical science publications and is not suitable for the wide range of disciplines. Google Scholar search does not provide a convenient way to download a database of publications. However, it is possible to use Google Scholar search to get a database with the other parsing script after some modifications. Script with modifications will be available soon at https://github.com/azarnyx/parse.py. Though, Google Scholar search output is restricted to 1000 articles, which might not be representative.
Interestingly, that nowadays such a search available directly on the Web Of Science search with Analyze Results button, however, it searches through all results rather than through 10000 most cited results as was used so it gave somehow different output:
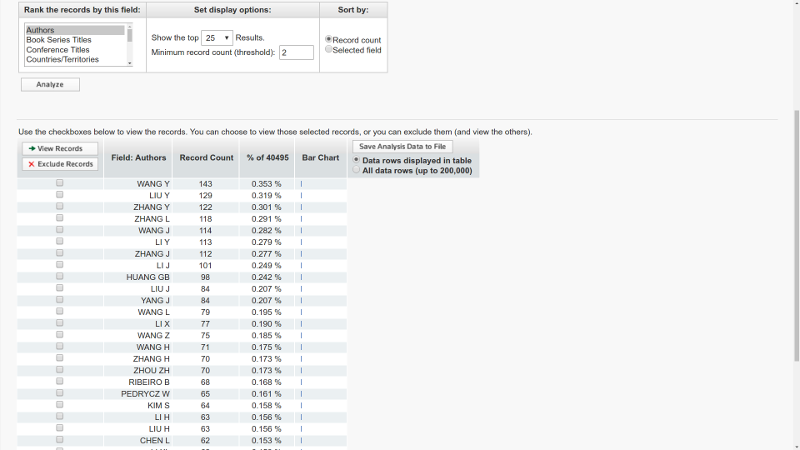
The WoS output still could be modified by an accurate setting of filters.
Recent Posts
- 12 Oct 2016 » Org-mode: beginners setup
- 18 Jul 2016 » Web of Science data parser